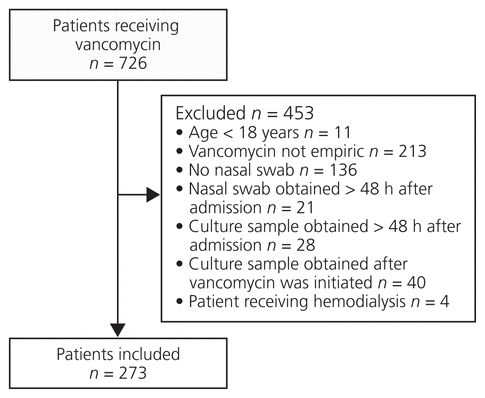
Figure 1 Identification of study sample from patients newly admitted to the Peter Lougheed Centre who were treated empirically with vancomycin. The exclusion criteria were applied sequentially in the order shown.
Josée Rioux, Jenny Edwards, Lauren Bresee, Adrian Abu-Ulba, Stephen Yu, Deonne Dersch-Mills, Ben WilsonABSTRACT
Background
Nasal-swab screening for methicillin-resistant Staphylococcus aureus (MRSA) has a quicker turnaround time than other bacterial culture methods, with results available within 24 h. Although MRSA nasal-swab screening is not intended to guide antimicrobial therapy, this method may give clinicians additional information for earlier tailoring of empiric antimicrobial agents.
Objective
To describe the diagnostic characteristics of nasal-swab screening in predicting MRSA infections in hospitalized patients receiving empiric treatment with IV vancomycin.
Methods
A retrospective observational chart review was conducted for newly admitted adult patients of the Peter Lougheed Centre in Calgary, Alberta, who were treated empirically with IV vancomycin from January to October 2015 and who underwent nasal-swab screening for MRSA. The diagnostic characteristics of nasal-swab screening were calculated in relation to corresponding culture results for samples collected on admission.
Results
For the 273 patients included in this study, nasal-swab screening for MRSA showed the following diagnostic characteristics in relation to bacterial culture results: sensitivity 58.3% (95% confidence interval [CI] 28.6%–83.5%), specificity 93.9% (95% CI 90.0%–96.3%), positive predictive value 30.4% (95% CI 14.1%–53.0%), negative predictive value 98.0% (95% CI 95.1%–99.3%), positive likelihood ratio 9.5 (95% CI 4.9–18.7), and negative likelihood ratio 0.4 (95% CI 0.2–0.9).
Conclusions
Given the high specificity of this rapid method, clinicians should ensure that patients who are receiving empiric treatment for MRSA infection and who have a positive result on nasal-swab screening continue to receive MRSA coverage until culture results are available. In addition, the high negative predictive value and positive likelihood ratio for nasal-swab screening in a low-prevalence setting suggest that a negative result significantly reduces the probability of MRSA infection. Although nasal-swab screening for MRSA is currently used for determining isolation precautions, this method also had utility in helping clinicians to predict the probability of MRSA infection and in guiding decisions about antimicrobial therapy.
KEYWORDS: methicillin-resistant Staphylococcus aureus, MRSA, nasal swabs, vancomycin, infection
RÉSUMÉ
Contexte
Le dépistage du Staphylococcus aureus résistant à la méthicilline (SARM) par écouvillonnage nasal procure des résultats d’examen plus promptement que les autres techniques de culture bactérienne, les résultats étant disponibles dans les 24 heures. Bien que les résultats du dépistage du SARM par écouvillonnage nasal ne soient pas destinés à guider le choix de traitement antimicrobien, cette technique peut fournir aux cliniciens des informations supplémentaires leur permettant de préciser plus rapidement les antibiothérapies empiriques adéquates.
Objectif
Présenter les caractéristiques diagnostiques du dépistage par écouvillonnage nasal comme outil servant à prédire les infections à SARM chez les patients hospitalisés qui reçoivent un traitement empirique de vancomycine par voie intraveineuse.
Méthodes
On a mené une analyse d’observation rétrospective au moyen des dossiers médicaux de patients adultes nouvellement admis au Peter Lougheed Centre à Calgary, en Alberta, ayant reçu un traitement empirique de vancomycine par voie intraveineuse entre janvier 2015 et octobre 2015 et ayant subi un dépistage du SARM par écouvillonnage nasal. Les caractéristiques diagnostiques du dépistage par écouvillonnage nasal ont été obtenues par comparaison avec les résultats de culture correspondants qui provenaient des échantillons recueillis à l’admission.
Résultats
Pour ce qui est des 273 patients retenus pour la présente étude, le dépistage du SARM par écouvillonnage nasal a affiché les caractéristiques diagnostiques suivantes comparativement aux résultats des cultures bactériennes : sensibilité de 58,3 % (intervalle de confiance [IC] à 95 % de 28,6 % à 83,5 %), spécificité de 93,9 % (IC à 95 % de 90,0 % à 96,3 %), valeur prédictive positive de 30,4 % (IC à 95 % de 14,1 % à 53,0 %), valeur prédictive négative de 98,0 % (IC à 95 % de 95,1 % à 99,3 %), rapport de vraisemblance positif de 9,5 (IC à 95 % de 4,9 à 18,7) et rapport de vraisemblance négatif de 0,4 (IC à 95 % de 0,2 à 0,9).
Conclusions
Compte tenu de la spécificité élevée de cette technique rapide, les cliniciens devraient s’assurer que les patients qui reçoivent un traitement empirique pour une infection à SARM et dont le résultat du dépistage du SARM par écouvillonnage nasal se révèle positif continuent à être traités contre le SARM jusqu’à l’obtention des résultats de culture. De plus, la valeur prédictive négative élevée et le rapport de vraisemblance positif élevé associés au dépistage par écouvillonnage nasal dans un contexte de faible prévalence suggèrent qu’un résultat négatif réduit de façon significative les probabilités d’infection à SARM. Enfin, bien que le dépistage du SARM par écouvillonnage nasal soit présentement utilisé pour déterminer les précautions à prendre concernant l’isolation, ce type d’analyse avait aussi le potentiel d’aider les cliniciens à prévoir les probabilités d’infection à SARM et de guider leur choix quant à l’antibiothérapie.
MOTS CLÉS: Staphylococcus aureus résistant à la méthicilline, SARM, écouvillonnage nasal, vancomycine, infection
Infections due to methicillin-resistant Staphylococcus aureus (MRSA) are a growing concern for the Canadian health care system.1,2 Alberta-specific data show that the incidence and prevalence of MRSA infection have been steady over the past few years: as of December 2015, the infection rate remained at 0.35 cases per 10 000 patient-days.3 MRSA infections result in increases in morbidity, length of hospital stay, and cost of treatment.1 Infectious disease guidelines recommend empiric IV vancomycin therapy for suspected MRSA infections.4–6 Culture and sensitivity results are subsequently used to tailor the antimicrobial therapy.4,5 However, negative culture results may take up to 120 h, and during this time clinicians are often hesitant to de-escalate MRSA coverage.7 Given that unnecessary antimicrobial use propagates antimicrobial resistance, it is important to narrow antimicrobial coverage as quickly as possible.
Patients with MRSA colonization are potential reservoirs for transmission and are themselves at higher risk of acquiring MRSA-associated infections.8–12 Nasal-swab screening is currently used at the study centre to identify MRSA carriers and to direct appropriate isolation of colonized patients.13 According to the local institutional policy for screening on admission, nasal swabbing is performed for patients with recognized risk factors for MRSA and for patients admitted to units where high MRSA risk has been identified, such as intensive care.13 Nasal swabs are usually obtained on admission by nursing staff, and the screening has a quicker turnaround time than other bacterial culture methods, with results available within 24 h at the study centre.14 Although MRSA nasal-swab screening is not intended to guide antimicrobial therapy, this method may give clinicians additional information to help in earlier tailoring of empiric antimicrobials.
The current literature illustrates a wide range of diagnostic characteristics for MRSA nasal-swab screening, with the majority of studies showing lower sensitivity (50%–88%) and higher specificity (83%–100%).15–20 The populations studied have included patients with various types of pneumonia, skin and soft-tissue infections, or multiple sites of potential infection, as well as patients admitted to the intensive care unit.15–20 Furthermore, the current literature15–20 focuses on patients with documented MRSA infection or patients at high risk for nosocomial infection, with patients being followed to determine whether they developed MRSA infection. To our knowledge, no studies have examined consecutive patients presenting to hospital who were started on empiric IV vancomycin therapy.
The primary objective of this study was to describe the diagnostic characteristics of MRSA nasal-swab screening in predicting culture-proven MRSA infection in newly admitted patients treated empirically with IV vancomycin. The secondary objectives were to describe the impact of admission to hospital within the prior 3 months on these diagnostic characteristics and to describe the diagnostic characteristics of MRSA nasal-swab screening according to bacterial culture site. This information may assist clinicians in early tailoring of empiric anti-MRSA therapy. Earlier discontinuation of anti-MRSA therapies could lead to reductions in resistance, adverse events, therapeutic drug monitoring, and overall costs.
A retrospective observational chart review was conducted of surveillance MRSA nasal-swab screening and corresponding culture results for clinical isolates of blood, sputum, wound, bronchoalveolar lavage (BAL), and endotracheal tube aspirate collected on admission from patients empirically treated with IV vancomycin. Working backward from October 2015, patients were included until the desired sample size was reached. The study was conducted at the Peter Lougheed Centre, a tertiary care centre in Calgary, Alberta, with approximately 600 beds, which services Calgary and surrounding areas.
A list of patients receiving IV vancomycin was pulled from the site’s inpatient medication dispensing program. Data were collected from a computerized charting database that includes patients’ demographic characteristics, clinicians’ orders, electronic medication administration records, and laboratory results.
Data collected consisted of patients’ demographic characteristics, admission date and time, admitting service, and MRSA nasal-swab and bacterial culture results (blood, sputum, wound, BAL, and endotracheal tube aspirate). The documented date and time of administration of the first dose of vancomycin and whether or not the patient had been admitted to hospital within the prior 3 months were also collected.
Patients eligible for inclusion were those 18 years of age or older who had been admitted to the Peter Lougheed Centre and had been initiated on empiric IV vancomycin (i.e., first dose administered within 48 h of admission). Patients had to have a documented MRSA nasal swab and culture of samples from at least one of the following sites, drawn within 48 h of admission: blood, sputum, wound, BAL, and/or endotracheal tube.
Patients with bacterial culture samples collected after the first IV dose of vancomycin were excluded. In addition, patients on long-term dialysis therapy were excluded because of their increased risk of non-MRSA infections requiring vancomycin therapy (e.g., line infections with coagulase-negative Staphylococcus). Finally, a given patient could be included in the study only once (even if there was more than one admission), and each patient could have data from only one MRSA nasal swab included. However, a patient could have culture data from 2 or more sites included; if the result for any of the multiple cultures was positive, the patient was considered culture-positive in the primary outcome analysis.
The required sample size was calculated using precision calculations based on different sensitivity and specificity estimates found in the current literature,15–20 and the largest sample size was chosen to ensure adequate precision for the study. The precision calculation was based on a 95% confidence interval (CI), an infinite population size, and a precision of 5%. Based on these parameters, a sample of 273 patients was required.
Descriptive statistics, including proportions for categorical variables and the mean and standard deviation for continuous variables, were used to report study group characteristics. Sensitivity, specificity, positive predictive value, negative predictive value, and likelihood ratios for the MRSA nasal-swab results in predicting MRSA infection were calculated (with 95% CIs). The same diagnostic characteristics were calculated for the secondary outcomes.
The study was approved by the Health Research Ethics Board of the Alberta Community Health Committee. A waiver of consent was granted because of the retrospective nature of the study.
A total of 726 patients were screened, and 453 of these individuals were excluded. The exclusion criteria were assessed sequentially in the order shown in Figure 1. The 273 patients included in the analysis were mostly male (176 [64.5%]), and the mean age was 55.8 years (standard deviation 17.7) (Table 1). The majority of patients were admitted under the care of internal medicine (146 [53.5%]) and hospitalist (69 [25.3%]) teams, and most did not have a hospital admission in the prior 3 months (200 [73.3%]). The most common bacterial culture site (based on 334 samples cultured) was blood (266 [79.6%]), followed by wound (36 [10.8%]). Twelve patients had a total of 14 positive MRSA culture results (7 from blood, 6 from wound, and 1 from BAL), and 23 positive MRSA nasal-swab results were identified (Table 2). Details of the culture results are shown in Tables 2 and 3.
|
|
||
|
Figure 1 Identification of study sample from patients newly admitted to the Peter Lougheed Centre who were treated empirically with vancomycin. The exclusion criteria were applied sequentially in the order shown. |
||
Table 1 Patient Characteristics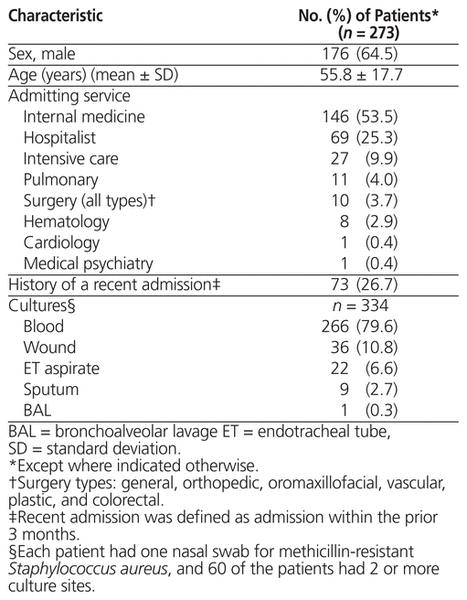
Table 2 MRSA Culture and Nasal Swabs Results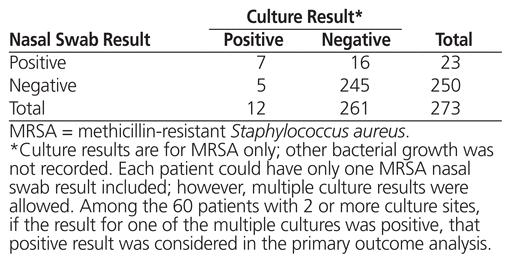
Table 3 Results by Culture Type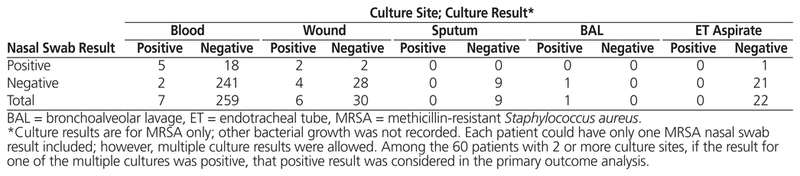
Nasal-swab screening for MRSA had the following diagnostic characteristics in relation to bacterial culture results: sensitivity 58.3% (95% CI 28.6%–83.5%), specificity 93.9% (95% CI 90.0%–96.3%), positive predictive value 30.4% (95% CI 14.1%–53.0%), negative predictive value 98.0% (95% CI 95.1%–99.3%), positive likelihood ratio 9.5 (95% CI 4.9–18.7), and negative likelihood ratio 0.4 (95% CI 0.2–0.9).
For the secondary objective, a recent admission resulted in similar specificity (91.3% [95% CI 81.4%–96.4%] versus 94.8% [95% CI 90.4%–97.3%]) and higher sensitivity (75.0% [95% CI 21.9%–98.7%] versus 50.0% [95% CI 17.5%–82.6%]), relative to patients without a recent admission. As for individual culture sites, blood and wound cultures were found to have high specificity, about 93%, with lower sensitivities. Complete results are shown in Table 4.
Table 4 Diagnostic Characteristics of Nasal Swabbing for MRSA Infection in Newly Admitted Patients Treated Empirically with IV Vancomycin*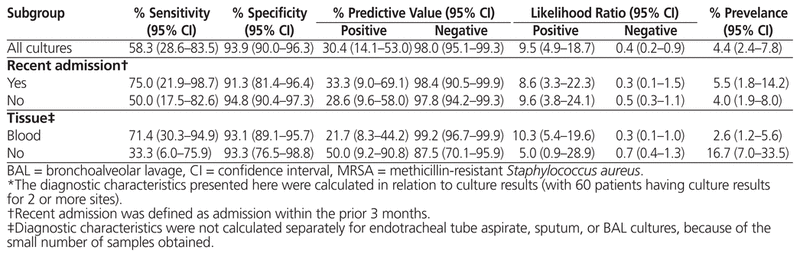
MRSA infections are associated with increased morbidity, mortality, and health care costs.1 Infectious disease guidelines recommend empiric antimicrobial coverage for MRSA when this type of infection is suspected; however, it is just as crucial to identify situations when MRSA coverage is not needed.
In this study, MRSA nasal-swab screening was associated with a negative predictive value of 98.0%. This high value suggests that the probability of MRSA infection in swab-negative patients is quite low and that MRSA infection in these patients would be unlikely. However, the relatively low prevalence of MRSA infection in the study population (4.4% [95% CI 2.4%–7.8%]) was a major contributor to the negative predictive value. For this reason, the negative predictive value suggests that, in appropriate clinical situations (including a population with low MRSA prevalence, similar to this study’s population), a negative MRSA swab result could be considered in the clinician’s decision to discontinue empiric vancomycin earlier.
The high specificity value for MRSA nasal-swab screening (93.9%) suggests that the likelihood of false-positive results is low and that MRSA nasal swabbing is useful for ruling in MRSA infection. As a result, for patients who are receiving empiric treatment for MRSA infection and who have a positive result on MRSA nasal swabbing, clinicians should ensure that MRSA coverage is continued until culture results are confirmed.
The low sensitivity value in this study (58.3%) indicates that nasal-swab screening has a high potential for false-negative results. There were 7 false-negative nasal-swab results among 5 patients in this study, corresponding to positive culture results for 2 blood samples, 4 wound samples, and 1 BAL sample. These false-negative nasal-swab results are important, given the risks associated with premature narrowing of antimicrobial therapy for MRSA-infected patients. Because of the frequency of false negatives, early discontinuation of anti-MRSA therapy should not be based solely upon a negative MRSA nasal-swab result and should be correlated with clinical judgment.
The positive likelihood value of 9.5 suggests that the post-test probability of MRSA infection in the presence of a positive MRSA nasal-swab result is increased by approximately 45%.21 The post-test probability can vary significantly according to the patient population being observed, and knowing the pre-test probability is key.
As for the secondary objectives, it was noted that recent admission had minimal impact on specificity (94.8% for those without recent admission versus 91.3% for those with recent admission) but increased the sensitivity (50.0% versus 75.0%, respectively). This difference is likely attributable to chance, resulting from the small number of samples cultured after stratification by admission history, and likely does not reflect a clinically significant increase in sensitivity.
Blood cultures made up the majority of the sample size, and generalizability of these results is therefore more likely limited to bloodstream infections and less applicable to wound infections. Unfortunately, there were too few individual endotracheal tube aspirate, sputum, and BAL samples to analyze. However, studies including respiratory samples15,17,18 have reported sensitivity and specificity similar to those found in the current study.
These results follow the same pattern—higher specificity than sensitivity—as findings from other studies involving MRSA nasal swabbing as single-site screening. Reported values for sensitivity and specificity of MRSA nasal-swab screening alone for predicting MRSA infection have ranged from 50.0% to 88.0% and from 83.0% to 100.0%, respectively.15–20 The comparability of these results is limited by the different nasal-swab assays used at each site. Many studies used polymerase chain reaction, which has been shown to increase sensitivity22 relative to the chromogenic medium used at the current study site.
In terms of local data, one Canadian study found similar diagnostic characteristics, except for a higher sensitivity of 91.0%.23 This difference in sensitivity is likely attributable to the use of a combination of nasal and rectal swabs, as well as selection of patients with culture-proven S. aureus infection.
Few studies have looked at the utility of MRSA swab–guided antimicrobial therapy in terms of clinical outcomes. One study of patients with suspected hospital-acquired pneumonia found that in the absence of respiratory culture, clinical outcomes were no worse when MRSA nasal- and throat-swab screening was used to guide vancomycin therapy.24 In addition to nasal and throat swabbing, this study also used a pulmonary infection severity scale.24 Use of a combination of swabbing sites and a clinical severity scale to guide the antimicrobial therapy likely reflects the reality of practice, with clinicians basing clinical decisions on multiple factors and not only the nasal-swab result in isolation.
A limitation of this study was the relatively small sample size. However, the required sample size was determined using the wide range of published diagnostic characteristics for MRSA nasal-swab screening, and should represent a sufficient number to provide an accurate estimate.
In addition, the assessment of a single swab site (the nasal passages) may have lowered the sensitivity relative to using a combination of results from MRSA swabs of different sites.25,26 Although both nasal and rectal swabbing are routinely performed at the study centre, nasal-swab results are reported within 24 h, whereas rectal-swab results take 4 days. The clinical advantage of identifying MRSA earlier than with other microbiological culture methods would be lost if rectal swabs were included. Because of the retrospective design of the study, we were unable to account for the variability in quality of nasal-swab collections. Patients who received treatment with MRSA-active antimicrobials outside of the study site were also unknown, as we did not have access to medication information that was not documented in the hospital’s charting system.
The strength of this study lies in the study population. Because patients were selected early in the admission process, the population reflects patients in whom early adjustment of empiric antimicrobial therapy may be reasonable. Furthermore, the study’s objectives are of high clinical relevance and are applicable across other practice sites. This study has also added to the scarce Canadian data available on the subject.
As a result of the high specificity of MRSA nasal-swab screening, for patients who are receiving empiric treatment for MRSA infection and are found to have a positive MRSA nasal-swab result, clinicians should ensure they continue to receive MRSA coverage until culture results are available. Additionally, the high negative predictive value and high positive likelihood ratio of MRSA nasal-swab screening in a low-prevalence setting suggest that a negative result on MRSA nasal-swab screening significantly reduces the probability of MRSA infection. Although MRSA nasal-swab screening is currently used for determining isolation precautions, this method also had utility in helping clinicians to better predict a patient’s probability of MRSA infection and in guiding antimicrobial decisions.
1 Goetghebeur M, Landry PA, Han D, Vicente C. Methicillin-resistant Staphylococcus aureus: a public health issue with economic consequences. Can J Infect Dis Med Microbiol. 2007;18(1):27–34.
2 Results of the surveillance of methicillin resistant Staphylococcus aureus — from 1995 to 2009 — a project of the Canadian Nosocomial Infection Surveillance Program (CNISP). Ottawa (ON): Public Health Agency of Canada; 2009.
3 Antimicrobial resistant organisms: MRSA and VRE 3rd quarter report: October – December 2015. Alberta: Alberta Health Services/Covenant Health, Infection Prevention and Control; 2016 Feb.
4 Interim guidelines for the management of community-associated methicillin-resistant Staphylococcus aureus infections in primary care. Vancouver (BC): BC Centre for Disease Control; 2006.
5 Rotstein C, Evans G, Born A, Grossman R, Light RB, Magder S, et al. Clinical practice guidelines for hospital-acquired pneumonia and ventilator-associated pneumonia in adults. Can J Infect Dis Med Microbiol. 2008;19(1):19–53.
6 Achiam CC, Fernandes CMB, McLeod SL, Salvadori MI, John M, Seabrook JA, et al. Methicillin-resistant Staphylococcus aureus in skin and soft tissue infections presenting to the emergency department of a Canadian academic health care center. Eur J Emerg Med. 2011;18(1):2–8.
7 Bacterial culture, aerobic culture only source listing. Calgary (AB): Calgary Laboratory Services; 2013 [cited 2015 Jul 24]. Available from: www.calgarylabservices.com/lab-services-guide/microbiology/Test/Tests/BacterialCulture.htm
8 Kluytmans JN, Belkum AV, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Am Soc Microbiol. 1997;10(3):505–20.
9 Davis K, Stewart JJ, Crouch HK, Florez CE, Hospenthal DR. Methicillin-resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clin Infect Dis. 2004;39(6): 776–82.
10 Ramirez MC, Marchessault M, Govednik-Horny C, Jupiter D, Papaconstantinou HT. The impact of MRSA colonization on surgical site infection following major gastrointestinal surgery. J Gastrointest Surg. 2013;17(1):144–52.
11 Niven DJ, Laupland KB, Gregson DB, Church DL. Epidemiology of Staphylococcus aureus nasal colonization and influence on outcome in the critically ill. J Crit Care. 2009;24(4):583–9.
12 Sim BLH, McBryde E, Street AC, Marshall C. Multiple site surveillance cultures as a predictor of methicillin-resistant Staphylococcus aureus infections. Infect Control Hosp Epidemiol. 2013;34(8):818–24.
13 Calgary Zone methicillin-resistant Staphylococcus aureus/vancomycin-resistant enterococci admission screening policy. Calgary (AB): Alberta Health Services, Infection Prevention and Control; 2013.
14 Methicillin resistant Staphylococcus aureus (MRSA) – nose, nasopharyngeal [test description]. Calgary (AB): Calgary Laboratory Services; 2013. Available from: www.calgarylabservices.com/lab-services-guide/microbiology/test/tests/MRSA-Nose.htm
15 Dangerfield B, Chung A, Webb B, Seville MT. Predictive value of methicillin-resistant Staphylococcus aureus (MRSA) nasal swab PCR assay for MRSA pneumonia. Antimicrob Agents Chemother. 2014;58(2):859–64.

16 Hennessy SA, Shah PM, Guidry CA, Davies SW, Hranjec T, Sawyer RG. Can nasal methicillin-resistant Staphylococcus aureus screening be used to avoid empiric vancomycin use in intra-abdominal infection? Surg Infect (Larchmt). 2015;16(4):396–400.
17 Langsjoen J, Brady C, Obenauf E, Kellie S. Nasal screening is useful in excluding methicillin-resistant Staphylococcus aureus in ventilator-associated pneumonia. Am J Infect Control. 2014;42(9):1014–5.
18 Tilahun B, Faust AC, McCorstin P, Ortegon A. Nasal colonization and lower respiratory tract infections with methicillin-resistant Staphylococcus aureus. Am J Crit Care. 2015;24(1):8–12.
19 Robicsek A, Suseno M, Beaumont JL, Thomson RB, Peterson LR. Prediction of methicillin-resistant Staphylococcus aureus involvement in disease sites by concomitant nasal sampling. J Clin Microbiol. 2008;46(2):588–92.

20 Schleyer AM, Jarman KM, Chan JD, Dellit TH. Role of nasal methicillin-resistant Staphylococcus aureus screening in the management of skin and soft tissue infections. Am J Infect Control. 2010;38(8):657–9.
21 McGee S. Simplifying likelihood ratios. J Gen Intern Med. 2002;17(8):647–50.
22 Byrnes MC, Adegboyega T, Riggle A, Chipman J, Beilman G, Reicks P, et al. Nasal swabs collected routinely to screen for colonization by methicillin-resistant Staphylococcus aureus in intensive care units are a sensitive screening test for the organism in clinical cultures. Surg Infect (Larchmt). 2010;11(6):511–5.
23 MacFadden DR, Elligsen M, Robicsek A, Ricciuto DR, Daneman N. Utility of prior screening for methicillin-resistant Staphylococcus aureus in predicting resistance of S. aureus infections. CMAJ. 2013;185(15):E725–30.

24 Boyce JM, Pop OF, Abreu-Lanfranco O, Hung WY, Fisher A, Karjoo A, et al. A trial of discontinuation of empiric vancomycin therapy in patients with suspected methicillin-resistant Staphylococcus aureus health care-associated pneumonia. Antimicrob Agents Chemother. 2013;57(3):1163–8.

25 Matheson A, Christie P, Stari T, Kavanagh K, Gould IM, Masterton R, et al. Nasal swab screening for methicillin-resistant Staphylococcus aureus—how well does it perform? A cross-sectional study. Infect Control Hosp Epidemiol. 2012;33(8):803–8.
26 Senn L, Basset P, Nahimana I, Zanetti G, Blanc DS. Which anatomical sites should be sampled for screening of methicillin-resistant Staphylococcus aureus carriage by culture or by rapid PCR test? Clin Microbiol Infect. 2012;18(2): E31–3.
Competing interests: None declared. ( Return to Text )
Funding: None received. ( Return to Text )
The authors would like to thank Pharmacy Services – Alberta Health Services for supporting staff members who worked on this study and for allowing data collection in the Peter Lougheed Centre.
Canadian Journal of Hospital Pharmacy, VOLUME 70, NUMBER 2, March-April 2017