Identification of Staphylococci by Polymerase Chain Reaction Directly from a Positive Blood Culture and Effect on Patient Care
Molly Wade-Cummings, Jonathan F Mailman, Michelle L Degelman, Casey Phillips, Jason R Vanstone
ABSTRACT
Background
As one of the most common bloodstream infections worldwide, Staphylococcus aureus bacteremia places a major burden on health care. Implementation of a rapid, genetic-based diagnostic test may have important implications in the clinical management of patients with S. aureus bacteremia.
Objectives
The primary objective was to assess concordance between testing based on polymerase chain reaction (PCR) and the current gold standard, culture and sensitivity testing; the secondary objective was to assess the impact of this technology on patient care.
Methods
A pre–post intervention retrospective chart review was used to document the hospital course of patients with a diagnosis of S. aureus bacteremia before and after implementation of the PCR-based diagnostic system. Laboratory results from all patient samples subjected to PCR-based analysis following implementation of this system were compared with culture and sensitivity data for the same samples to determine accuracy of the new system. In addition, time to optimal therapy for each patient was calculated as the interval between the initiation of empiric and terminal therapies. The appropriateness of antimicrobial treatment was characterized as guideline-concordant, nonconcordant with the guidelines, or nonconcordant yet still clinically appropriate.
Results
In total, 98 and 99 patients met the inclusion criteria before and after implementation of the PCR-based diagnostic system, respectively. PCR-based results displayed 99.8% concordance (440/441 total samples) with results from culture and sensitivity testing. The time to optimal therapy was significantly shorter after implementation, by a mean of 22.8 h (p < 0.001). Overall, 97% of empiric and 99% of terminal antimicrobial regimens were either guideline-concordant or clinically appropriate for treatment of S. aureus bacteremia; 3% of empiric and 1% of terminal antimicrobial regimens were nonconcordant with clinical guidelines without any explanation based on other clinical considerations.
Conclusions
The study findings support the utility of using a direct-from-positive-blood-culture PCR-based diagnostic tool as the primary method of identifying S. aureus bacteremia in patients, as well as the acceptance of and acting upon the new assay’s results by our local clinicians. PCR-based assays can help reduce the time to optimal terminal therapy for patients with bacteremia.
KEYWORDS: bacteremia, antimicrobial stewardship, polymerase chain reaction, diagnostic testing, GeneXpert
RÉSUMÉ
Contexte
La bactériémie à Staphylococcus aureus (BAC-SA), qui est l’une des infections du sang les plus répandues dans le monde, fait peser une lourde charge sur les soins de santé. La mise en place d’un test diagnostique génétique rapide pourrait avoir des retombées importantes sur la gestion clinique des patients présentant une BAC-SA.
Objectifs
L’objectif principal consistait à évaluer la concordance entre les tests basés sur la réaction en chaîne par polymérase (PCR) et le test de sensibilité et de culture, qui est la référence absolue actuelle; l’objectif secondaire consistait à évaluer l’impact de cette technologie sur les soins des patients.
Méthodes
Un examen rétrospectif des dossiers pré-et post-intervention a servi à décrire le séjour à l’hôpital des patients ayant reçu un diagnostic de BAC-SA avant et après la mise en place du système de diagnostic de la PCR. Les résultats de laboratoire de tous les échantillons des patients soumis à une analyse de la PCR à la suite de la mise en place de ce système ont été comparés avec les données relatives à la culture et à la sensibilité de ces mêmes échantillons afin de déterminer la précision du nouveau système. De plus, l’évaluation du délai d’atteinte du traitement optimal de chaque patient repose sur le calcul de l’intervalle entre le début des thérapies empiriques et terminales. La pertinence du traitement antimicrobien était caractérisée comme suit : concordance avec les lignes directrices, non-concordance avec les lignes directrices ou non-concordance mais encore approprié d’un point de vue clinique.
Résultats
Au total, 98 et 99 patients ont satisfait au critère d’inclusion respectivement avant et après la mise en place du système de diagnostic de la PCR. Les résultats basés sur la PCR affichaient une concordance de 99,8 % (440/441 échantillons au total) avec les résultats des tests de sensibilité et de culture. La diminution du délai d’atteinte du traitement optimal était importante après la mise en place du système, puisqu’elle atteignait en moyenne 22,8 h (p < 0,001). De manière générale, 97 % des régimes antimicrobiens empiriques et 99 % des régimes antimicrobiens terminaux concordaient avec les lignes directrices ou étaient cliniquement appropriés pour le traitement de la BAC-SA; 3 % des régimes antimicrobiens empiriques et 1 % des régimes antimicrobiens terminaux n’étaient pas conformes aux lignes directrices cliniques sans qu’aucune explication basée sur d’autres considérations cliniques n’ait été donnée.
Conclusions
Les résultats de l’étude confirment la nécessité d’utiliser un outil diagnostique basé sur la PCR directement de l’hémoculture positive en guise de méthode principale pour déterminer la présence de BAC-SA chez les patients ainsi que l’acceptation et l’utilisation des nouveaux résultats du test par nos cliniciens locaux. Les tests basés sur la PCR peuvent aider à réduire le délai d’attente du traitement optimal pour les patients atteints de BAC-SA.
MOTS CLÉS: bactériémie, gestion de l’utilisation des antimicrobiens, réaction en chaîne par polymérase, test de diagnostic, GeneXpert
INTRODUCTION
As one of the most common bloodstream infections worldwide, Staphylococcus aureus bacteremia places a major burden on health care.1 Despite existing as a commensal on the skin and in the nares, S. aureus in the bloodstream can result in an invasive disease, contributing to clinical illness and notoriously high mortality rates.2 With the current prevalence of methicillin-resistant S. aureus (MRSA) above 20% in the Regina area (unpublished laboratory data), individuals suspected of having S. aureus bacteremia are typically started on empiric therapy with broad-spectrum antibiotics to cover both MRSA and methicillin-susceptible S. aureus (MSSA) until microorganisms are identified and susceptibilities are available. Using conventional methods, it may take 48 h or longer to identify the microorganism and perform susceptibility testing after a positive blood culture result has been obtained.3,4 Not only does this lag period contribute to the potential for drug toxicities and antimicrobial resistance, but the length of time to optimal therapy has been shown to directly influence infection-related mortality and length of stay.2,5
In 2018, the Saskatchewan Health Authority – Regina Area began to employ a new method for identifying S. aureus bacteremia. Using the GeneXpert IV system, the Xpert MRSA/SA BC assay (Cepheid) allows identification of MRSA and MSSA using material obtained directly from a blood culture sample when gram-positive cocci in clusters have been identified. This system uses real-time polymerase chain reaction (PCR) to amplify and detect 3 distinct staphylococci genes: the gene for staphylococcal protein A (spa), which verifies the identity of S. aureus; a specific junction of the staphylococcal cassette chromosome (SCCmec); and the mecA gene for methicillin resistance. The detection of all 3 genes indicates an MRSA-positive sample, whereas the detection of spa alone or in conjunction with SCCmec indicates MSSA, and the absence of spa indicates other gram-positive cocci (for example, coagulase-negative staphylococci or micrococci).6 The GeneXpert system has the potential to allow clinicians to identify MRSA/MSSA from positive blood culture samples in under 1 h.7
Relative to traditional culture and sensitivity testing, the GeneXpert system has been shown to identify bacterial strains with high sensitivity and specificity, dramatically reduce time to optimal antibiotic therapy, and reduce overall use of empiric agents.8,9 The limit of detection has been reported as 600 colony-forming units (CFU)/mL for S. aureus and 800 CFU/mL for MRSA, comparable to direct culture methods but substantially higher than enrichment cultures used in laboratory settings.10,11 In the case of samples containing mixed bacterial species in quantities near the limit of detection, the risk of false or variable results increases; however, compared with reference culture, the sensitivity of GeneXpert for 792 specimens was reported as 98.1% for MRSA-positive samples, 99.6% for MRSA-negative samples, 99.6% for S. aureus–positive samples, and 99.5% for specimens negative for S. aureus. Additionally, testing of 101 different gram-positive, gram-negative, and yeast strains revealed 100% analytical specificity in which all results were reported as MRSA-negative and S. aureus–negative by the GeneXpert assay.10
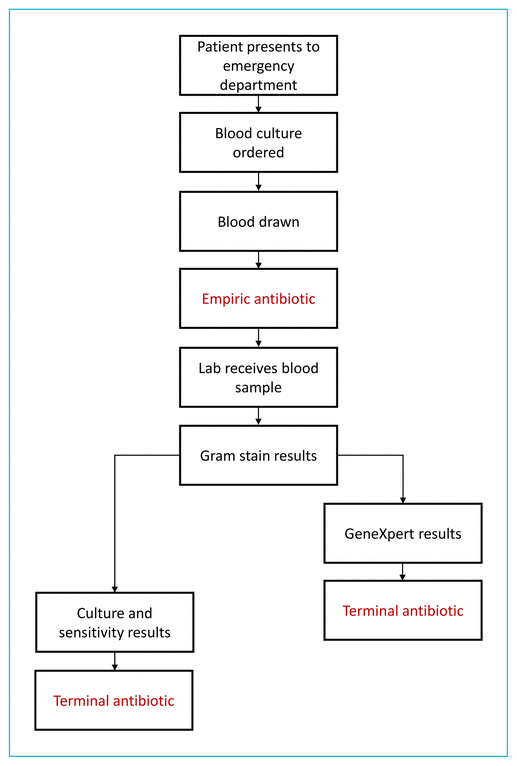
Although substantial evidence has shown the utility of PCR-based assays, few researchers have investigated their influence on patient care. We evaluated the patient care process from initial presentation to the emergency department to terminal antimicrobial treatment (Figure 1). The primary objective was to assess the accuracy of the GeneXpert system relative to the current gold standard of culture and sensitivity testing, and the secondary objective was to assess the impact of the GeneXpert system on patient care.
| |
|
|
|
FIGURE 1 The flow of care for patients with a diagnosis of Staphylococcus aureus bacteremia. After presentation to the emergency department, blood culture was ordered for patients with suspected bacteremia. Blood samples were drawn before initiation of an empiric antibiotic, sent to the laboratory, and subjected to Gram staining. Each patient’s terminal antimicrobial therapy was initiated either after culture and sensitivity testing (pre-implementation group) or after receipt of GeneXpert results (post-implementation group).
|
METHODS
This pre–post intervention retrospective cohort chart review involved patients in the Saskatchewan Health Authority – Regina Area admitted to either the Regina General Hospital or Pasqua Hospital or treated as outpatients in the emergency department of either hospital. The study population included patients 18 years of age or older who received treatment for S. aureus bacteremia. Admitted patients and emergency outpatients identified as having gram-positive cocci in clusters based on Gram staining of a cultured blood sample between October 17, 2017, and April 11, 2018 (pre-implementation of the GeneXpert system) and between April 12, 2018, and June 2, 2019 (post-implementation) were eligible for inclusion. Patients receiving outpatient hemodialysis were excluded because of the inaccessibility of their medical files. Randomly selected convenience samples of 99 patients before implementation and 98 patients after implementation were included.
Data for the following variables were collected: demographic and clinical characteristics (age, Sequential Organ Failure Assessment [SOFA] score,12 Acute Physiology and Chronic Health Evaluation [APACHE] II score,13 Charlson Comorbidity Index,14 MRSA risk factors [IV drug use, MRSA history, positive MRSA screening result, hospital admission or antimicrobial use in the past 90 days, overcrowded living conditions, chronic illnesses, indwelling devices or prostheses],15 primary source of infection,16 most responsible admission diagnosis, length of stay, discharge disposition, date and time of presentation to the emergency department, date and time of triage), blood culture information (date and time that blood cultures were ordered and samples were drawn and received by the laboratory, as well as date and time of reported results from Gram staining, GeneXpert analysis, and culture and sensitivity testing), and information on antimicrobial use (date and time of empiric and terminal antibiotic therapy, as well as appropriateness). An unpaired t test was used to identify statistically significant differences between the pre- and post-implementation groups in terms of mean age, length of stay, SOFA score, APACHE II score, Charlson comorbidity index, and time to optimal therapy. Categorical data (e.g., discharge disposition) were compared with a χ2 test. All information was captured using the online Research Electronic Database Capture (REDCap) tool.17,18
Notification of laboratory results was the same before and after implementation, whereby positive Gram stain results were immediately conveyed by telephone to the ordering ward, and organism identification and susceptibility reports were made available in the electronic health record. Laboratory results from all patient samples subjected to GeneXpert analysis (April 12, 2018, to June 2, 2019) were compared with corresponding culture and sensitivity data to determine concordance between methods.
The date and time of antimicrobial therapy were determined from administration times documented in the medication administration record of the patient’s medical chart. Empiric and terminal therapies were recorded as the first occurrence of the respective antimicrobials in the medication administration record. If antimicrobial therapy was terminated after laboratory results became available (e.g., if only 1 of 4 culture bottles had a positive result for gram-positive cocci and was ruled a contaminant), the date and time of terminal therapy was considered to be the final dose of empiric therapy documented in the medication administration record. If a patient remained on empiric therapy (e.g., tested positive for MRSA), terminal therapy was recorded as the date and time that either culture and sensitivity results became available (before implementation of the GeneXpert system) or GeneXpert results became available (after implementation). For each patient, time to optimal therapy was calculated as the interval between the initiation of empiric and terminal therapies and plotted on a control chart (X chart; QI Macros, KnowWhere International) for analysis of change signals in the data.
The appropriateness of antimicrobial treatment was characterized as guideline-concordant, nonconcordant, or nonconcordant yet still clinically appropriate.19 Guideline concordance was defined as the use of vancomycin, linezolid, or daptomycin as empiric therapy for patients with positive blood culture results and as terminal therapy for patients with MRSA; cloxacillin or cefazolin as terminal therapy for MSSA; and either discontinuation of antibiotic therapy or use of an optimal narrow-spectrum agent for other gram-positive cocci identified. Empiric treatment with cloxacillin or cefazolin for patients without other risk factors for MRSA (e.g., those with young age, few comorbidities, no previous hospital admissions or antimicrobial use) was considered guideline-nonconcordant yet clinically appropriate. Nonconcordance was defined as the use of sulfamethoxazole-trimethoprim for MRSA and empiric agents such as piperacillin-tazobactam, ertapenem, meropenem, or ceftriaxone for MRSA-negative infections in the absence of other risk factors or infections in other body sites. For cases in which therapy was deemed nonconcordant, a clinical pharmacist (J.F.M. or C.P.) reviewed patient comorbidities, the clinical picture, and MRSA risk factors to assess whether the treatment was clinically appropriate. For example, prolonged, broader-spectrum empiric treatment of patients with multiple comorbidities or sources of infection, despite narrower-spectrum therapies being available, would be considered nonconcordant yet clinically appropriate antimicrobial use.
This study was exempted from ethical review by the former Regina Qu’Appelle Health Region (RQHR) Research Ethics Board (REB-19-62) and was conducted in accordance with the ethical standards of the former RQHR Research Ethics Board, the Tri-Council Policy Statement: Ethical Conduct for Research Involving Humans, and the Helsinki Declaration.
RESULTS
Patient Characteristics
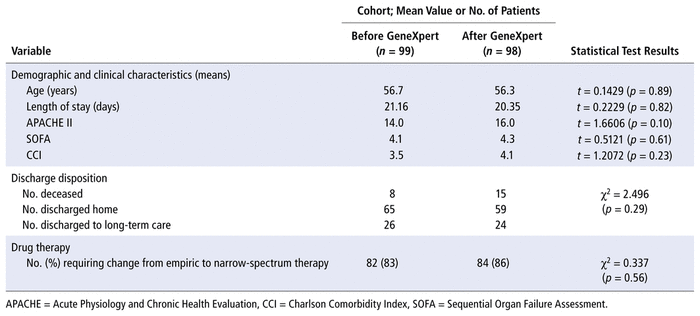
Of the 263 charts reviewed, 66 were excluded because the patients were not treated for S. aureus bacteremia (left against medical advice, discharged before antibiotic administration, or not receiving any antimicrobial therapy because a positive culture result was ruled as representing a contaminant), because they did not receive an initial antibiotic before blood culture results became available, or because blood culture samples were not tested by the GeneXpert system in the post-implementation phase. Patient demographic characteristics in the 2 groups are presented in Table 1. There were no significant differences between groups in average age, disposition, length of stay, SOFA score, APACHE II score, or Charlson comorbidity index.
TABLE 1 Characteristics of Cohorts before and after Implementation of the GeneXpert System
Laboratory Results
The ratio of MRSA to MSSA to other gram-positive cocci was similar before implementation of the GeneXpert system (17:38:44) and after implementation (14:26:58).
Concordance with Culture and Sensitivity Testing
For all samples tested, the GeneXpert system displayed 99.8% concordance (440/441) relative to culture and sensitivity testing. The assay falsely identified 1 sample as MRSA-positive, whereas further susceptibility testing showed that it was MSSA-positive.
Time to Optimal Therapy
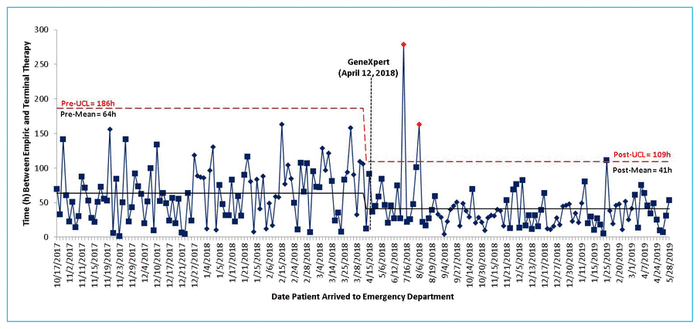
From the time of initial blood sampling, the mean intervals to reported Gram staining results, GeneXpert results, and final culture and sensitivity results were 23.1 h, 26.4 h, and 100.6 h, respectively. A statistically significant difference was found in the time to optimal therapy between groups: mean time 63.6 (standard deviation [SD] 122.4) h before implementation of the GeneXpert system and 40.8 (SD 68.5) h after implementation, a difference of 22.8 h (p < 0.001). The control chart analysis did not reveal any special-cause variation in the time to optimal antimicrobial therapy. However, a decrease in the upper 3-σ control limit was observed for patients treated after implementation of the GeneXpert system, which indicates less variability in the data points and more frequent observation of shorter lengths of time to optimal therapy relative to the pre-intervention group (Figure 2).
| |
|
|
|
FIGURE 2 Time (h) between administration of empiric and terminal therapy before (October 17, 2017, to April 11, 2018) and after (April 12, 2018, to June 2, 2019) implementation of the GeneXpert system, which occurred on April 12, 2018 (black dashed line). Data from patients in the post-implementation group showed a decrease in the upper 3-σ control limit (UCL, red dashed line), indicating less variability in the data, and a reduction in the mean time to optimal terminal therapy (solid black line). These results suggest that the GeneXpert system allowed initiation of optimal terminal therapy in a shorter period of time and more consistently, relative to patients in the pre-implementation group.
|
Antimicrobials
For the pre- and post-implementation groups combined, 97% (191/197) of empiric regimens and 99% (198/200) of terminal antimicrobial therapies were either guideline-concordant or clinically appropriate for the treatment of S. aureus bacteremia. Conversely, 3% (6/197) of empiric regimens and 1% (2/200) of terminal antimicrobial therapies were nonconcordant with clinical guidelines without any explanation based on other clinical considerations.
DISCUSSION
The utility of rapid PCR-based assays in the treatment of S. aureus bacteremia has been increasingly recognized in the past decade.20 When the clinical role of a tool such as the GeneXpert system is being considered, speed and accuracy are key determinants, in addition to cost, influence on workflow, and laboratory implementation. As a quality control measure, we compared the concordance between the GeneXpert system and culture and sensitivity results from the same blood samples with positive Gram staining results. Among 441 samples, only 1 instance of discordance was observed over a period of approximately 5 months after implementation. In that case, the causative organism was identified as MRSA by the GeneXpert system but displayed cloxacillin sensitivity; the culture and sensitivity results were regarded as the gold standard and were used to facilitate patient treatment. The risk of false-positive and false-negative test results is one limitation associated with the use of molecular assays. False detection of mecA in an empty staphylococcal cassette chromosome is a predominant cause of false positives, whereas borderline oxacillin resistance, resistance through alternative mechanisms (altered penicillin binding proteins), or the presence of the mecA homologue mecC contribute to false-negative results.10,21 Through distinct detection of mecA in addition to SCCmec and spa, the primers and probes of the GeneXpert system minimize the prevalence of false detection relative to other assays; however, this technology should continue to be re-evaluated as new bacterial variants emerge.6,22
The greater speed of detection of the GeneXpert system relative to conventional culture and sensitivity testing remains uncontested, yet the implications of this speed for patient care remain poorly defined. We evaluated the influence of the GeneXpert system on time to optimal antibiotic therapy. Because there were no statistically significant differences in age, SOFA score, APACHE II score, or Charlson comorbidity index between the patient groups, any significant differences in the study variables were attributed to use of the GeneXpert system. We found a significant reduction in the mean time to optimal therapy after implementation of the GeneXpert system, by 22.8 h. This indicates acceptance of diagnostic results from the new system by local clinicians and subsequent changes to antimicrobial therapy for patients. The importance of earlier microbiology results and shorter time to optimal treatment has been reported in numerous studies. For example, in their retrospective trial, Lodise and others5 found that delayed treatment was an independent predictor of infection-related mortality in patients with S. aureus bacteremia, whereas another retrospective study examining 684 cases of S. aureus bacteremia concluded that patients with the longest time to blood culture positivity had a 30-day mortality rate of 39%, compared with 17% for those with early detection.23 Our study was relatively small, and we did not observe any difference in mortality between groups; this may be an avenue for future research.
In terms of cost (where all costs are reported in Canadian dollars, unless otherwise noted, and are relevant to the date of reporting and the respective currency), workflow, and practicality of laboratory implementation, the feasibility of integrating the GeneXpert system into tertiary care is an important consideration. In 2010, the list prices for the PCR test and GeneXpert IV system were $65 USD and $35 000 USD, respectively.24 Total implementation costs at our centre were estimated at $33 320.60 per year, excluding the price of the GeneXpert instrumentation, which was already being utilized by our microbiology laboratory for other testing. Implementation costs included monthly quality control ($819.60 per year), annual proficiency testing ($400 per year), and the cost of patient testing based on the number of positive blood culture results in 2016 ($68.30 per patient for 470 patients with positive blood culture results, for a total estimated cost of $32 101 per year). We did not directly evaluate the potential cost savings associated with empiric antimicrobial usage before and after implementation of the GeneXpert system; however, a 10-day course of vancomycin is more expensive ($234–$856) than cloxacillin ($72–$134), cephalexin ($30–$50), and other narrow-spectrum antimicrobials.25 Previous studies have shown little difference in hospital costs accrued during the days of antimicrobial therapy with GeneXpert system use; however, one study demonstrated that mean hospital costs were $21 387 USD less, over a 4-month period, after implementation because of fewer patient-days spent in the intensive care unit (ICU) and reduced length of hospital stay.24 Similarly, GeneXpert technology was found to reduce the number of ICU isolation days by 44% relative to conventional culture methods, with an estimated cost savings of €121.76 per isolation day avoided.26,27
Despite the 20% rate of MRSA in our area, most patients (166/197) were identified as having infection with MSSA or infection caused by other gram-positive cocci. The high incidence of non–S. aureus infections (102/197) may be attributed to the clinical prevalence of coagulase-negative staphylococci. This group of organisms has become a major cause of nosocomial infection and, in addition to S. aureus, represents one of the most common blood culture isolates, whether due to true infection or contamination from the skin surface.28 From an antimicrobial stewardship standpoint, being able to rule out MRSA sooner in these patients may limit the use of empiric agents such as vancomycin, reducing the potential for adverse drug events and the development of resistant organisms such as vancomycin-resistant enterococci or vancomycin-resistant S. aureus.8,15,26 Additionally, earlier access to MRSA-negative results may enable practitioners to discharge patients to home with IV antimicrobial therapy and avoid calling them back for treatment once laboratory results are available, thereby decreasing the utilization of downstream resources. For the 31 patients with positive results for MRSA, the delay in confirmation of presence of this organism likely had little effect on outcomes, because empiric coverage with an antimicrobial having MRSA activity (such as vancomycin) is standard in the tertiary centres participating in this study. Faster identification of MRSA may have more utility in centres with lower community rates of MRSA, which do not typically initiate vancomycin for S. aureus bacteremia, enabling faster coverage and an increased potential for beneficial patient outcomes.29
The control chart (Figure 2) did not reveal any special-cause variation in the data. It was anticipated that there might be a downward shift (i.e., a reduction in the length of time to optimal therapy) after implementation of the GeneXpert system; however, inherent variability in individual data points resulted in lack of an obvious trend. We did observe a drop in overall mean time to optimal therapy. After implementation of the GeneXpert system, there was less variability in time to optimal therapy as the data points produced a smaller upper 3-σ control limit. This suggests that clinicians were more consistently able to provide optimal terminal antibiotic therapy sooner following implementation of the GeneXpert system.
There was no significant difference in length of stay between the pre- and post-implementation groups. One of the earlier studies of this platform, conducted by Bauer and others24 and published in 2010, demonstrated the power of this tool to reduce patient length of stay by 6.2 days on average, a difference that was associated with a noticeable cost reduction because patients spent less time in the ICU. The difference in outcomes between studies may be attributed to the study populations, given that the majority of patients in the earlier study were from the ICU (66% pre- and 67% post-implementation), whereas our population consisted largely of emergency outpatients. Additional factors may include advances in medical care over time, given that the studies were conducted nearly a decade apart, or the previously mentioned inherent variability in patient data resulting in no significant difference in patient length of stay between our groups.
The clinical management of S. aureus bacteremia was found to be acceptable, as nearly all antimicrobial regimens were either concordant with guidelines or nonconcordant but still clinically appropriate. There did appear to be a lag associated with terminal antibiotic administration, even with use of the GeneXpert system: on average, results were available within 26.4 h, yet mean time to optimal terminal therapy was 40.8 h. This delay may have been influenced by factors such as clinical decision-making, dosing intervals for antimicrobial drugs, or potential discrepancies in charting or record keeping in relation to the time of actual drug administration; therefore, it is difficult to pinpoint clinical interventions that might shorten this time frame.
The limitations of this study include the retrospective nature of chart reviews, which has been described as increasing susceptibility to bias in data selection and leading to greater difficulty in establishing causal relationships than prospective studies.30 Additionally, this was a relatively small, single-centre study. With more data, we might see a more significant difference in variables, such as discharge disposition. The strengths of this study include the random selection of patients from a relatively large pool, which reduces the potential for selection bias, as well as the combination of analyses for quality improvement and parametric and nonparametric statistical methods. This approach allowed us to analyze data from individual samples to identify trends, as well as to compare means between groups.
CONCLUSION
The implementation of a new, rapid diagnostic technology for the identification of S. aureus bacteremia is a practical step in the clinical management of patients. The GeneXpert system displayed a high level of concordance with the results of conventional culture and sensitivity testing and significantly decreased the time to initiation of terminal antimicrobial therapy. PCR-based assays play an important role at the frontier of antimicrobial stewardship by enabling faster diagnosis and a reduction in the use of broad-spectrum agents, which may help combat previously reported high mortality rates associated with S. aureus bacteremia and the progression of antimicrobial resistance.
References
1 Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28(3):603–61.
Crossref PubMed PMC
2 Youssef D, Molony K. Staphylococcus aureus bacteremia in adults. In: Enany S, Alexander LEC, editors. Frontiers in Staphylococcus aureus. IntechOpen; 2017. p. 117–31.
3 Brown DFJ, Edwards DI, Hawkey PM, Morrison D, Ridgway GL, Towner KJ, et al. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). J Antimicrob Chemother. 2005;56(6):1000–18.
Crossref PubMed
4 The RQHR microbiology test compendium. Regina Qu’Appelle Health Region; 2015.
5 Lodise TP, McKinnon PS, Swiderski L, Rybak MJ. Outcomes analysis of delayed antibiotic treatment for hospital-acquired Staphylococcus aureus bacteremia. Clin Infect Dis. 2003;36(11):1418–23.
Crossref PubMed
6 Buchan BW, Allen S, Burnham CAD, Tekippe EM, Davis T, Levi M, et al. Comparison of the next-generation Xpert MRSA/SA BC assay and the GeneOhm StaphSR assay to routine culture for identification of Staphylococcus aureus and methicillin-resistant S. aureus in positive-blood-culture broths. J Clin Microbiol. 2015;53(3):804–9.
Crossref PMC
7 Wolk DM, Struelens MJ, Pancholi P, Davis T, Della-Latta P, Fuller D, et al. Rapid detection of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in wound specimens and blood cultures: multicenter preclinical evaluation of the Cepheid Xpert MRSA/SA skin and soft tissue and blood culture assays. J Clin Microbiol. 2009;47(3):823–6.
Crossref PubMed PMC
8 Davies J, Gordon CL, Tong SYC, Baird RW, Davis JS. Impact of results of a rapid Staphylococcus aureus diagnostic test on prescribing of antibiotics for patients with clustered gram-positive cocci in blood cultures. J Clin Microbiol. 2012;50(6):2056–8.
Crossref PubMed PMC
9 Ben-Zvi H, Drozdinsky G, Kushnir S, Avni T, Scheuerman O, Bisahra J, et al. Influence of GeneXpert MRSA/SA test implementation on clinical outcomes of Staphylococcus aureus bacteremia – a before-after retrospective study. Diagn Microbiol Infect Dis. 2019;93(2):120–4.
Crossref
10 Xpert MRSA-SA BC [package insert]. Cepheid; 2008.
11 Rossney AS, Herra CM, Brennan GI, Morgan PM, O’Connell B. Evaluation of the Xpert methicillin-resistant Staphylococcus aureus (MRSA) assay using the GeneXpert real-time PCR platform for rapid detection of MRSA from screening specimens. J Clin Microbiol. 2008;46(10):3285–90.
Crossref PubMed PMC
12 Vincent JL, Moreno R, Takala J, Willatts S, De Mendonça A, Bruining H, et al.; Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. Intensive Care Med. 1996;22(7):707–10.
Crossref PubMed
13 Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13(10):818–29.
Crossref PubMed
14 Charlson ME, Pompei P, Ales KL, MacKenzie R. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–83.
Crossref PubMed
15 Wooten DA, Winston LG. Risk factors for methicillin-resistant Staphylococcus aureus in patients with community-onset and hospital-onset pneumonia. Respir Med. 2013;107(8):1266–70.
Crossref PubMed
16 Bloodstream infections (BSI) surveillance protocol. Alberta Health Services; 2019.
17 Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–81.
Crossref PMC
18 Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95:103208.
Crossref PubMed PMC
19 Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, et al. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Infect Dis Clin Pract. 2011;52(3):18–55.
Crossref
20 Kothari A, Morgan M, Haake DA. Emerging technologies for rapid identification of bloodstream pathogens. Clin Infect Dis. 2014;59(2):272–8.
Crossref PubMed PMC
21 Blanc DS, Basset P, Nahimana-Tessemo I, Jaton K, Greub G, Zanetti G. High proportion of wrongly identified methicillin-resistant Staphylococcus aureus carriers by use of a rapid commercial PCR assay due to presence of staphylococcal cassette chromosome element lacking the mecA gene. J Clin Microbiol. 2011;49(2):722–4.
Crossref PMC
22 Bakthavatchalam YD, Nabarro LEB, Veeraraghavan B. Evolving rapid methicillin-resistant Staphylococcus aureus detection: cover all the bases. J Glob Infect Dis. 2017;9(1):18–22.
Crossref PubMed PMC
23 Kim J, Gregson DB, Ross T, Laupland KB. Time to blood culture positivity in Staphylococcus aureus bacteremia: association with 30-day mortality. J Infect. 2010;61(3):197–204.
Crossref PubMed
24 Bauer KA, West JE, Balada-Llasat JM, Pancholi P, Stevenson KB, Goff DA. An antimicrobial stewardship program’s impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin Infect Dis. 2010;51(9):1074–80.
Crossref PubMed
25 Antibiotics and common infections. RxFiles Canada; 2016 Oct.
26 Wassenberg M, Kluytmans J, Erdkamp S, Bosboom R, Buiting A, van Elzakker E, et al. Costs and benefits of rapid screening of methicillin-resistant Staphylococcus aureus carriage in intensive care units: a prospective multicenter study. Crit Care. 2012;16(1):Article R22.
Crossref PubMed PMC
27 Wassenberg MWM, Kluytmans JAJW, Box ATA, Bosboom RW, Buiting AGM, van Elzakker EPM, et al. Rapid screening of methicillin-resistant Staphylococcus aureus using PCR and chromogenic agar: a prospective study to evaluate costs and effects. Clin Microbiol Infect. 2010;16(12):1754–61.
Crossref PubMed
28 Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27(4):870–926.
Crossref PubMed PMC
29 Paul M, Kariv G, Goldberg E, Raskin M, Shaked H, Hazzan R, et al. Importance of appropriate empirical antibiotic therapy for methicillin-resistant Staphylococcus aureus bacteraemia. J Antimicrob Chemother. 2010;65(12):2658–65.
Crossref PubMed
30 Ward RA, Brier ME. Retrospective analyses of large medical databases: what do they tell us? J Am Soc Nephrol. 1999;10(2):429–32.
PubMed
Molly Wade-Cummings, BSc, is with the College of Pharmacy and Nutrition, University of Saskatchewan, Saskatoon, Saskatchewan
Jonathan F Mailman, BSc(Pharm), ACPR, PharmD, CD, is with the Department of Pharmacy Services, Saskatchewan Health Authority, Regina, Saskatchewan, and the College of Medicine, University of Saskatchewan, Saskatoon, Saskatchewan
Michelle L Degelman, MSc, is with Stewardship and Clinical Appropriateness, Saskatchewan Health Authority, Regina, Saskatchewan
Casey Phillips, BSP, PharmD, is with the Antimicrobial Stewardship Program, Saskatchewan Health Authority, Regina, Saskatchewan
Jason R Vanstone, PhD, is with Stewardship and Clinical Appropriateness, Saskatchewan Health Authority, Regina, Saskatchewan
Competing interests: None declared. (
Return to Text
)
Address correspondence to: Dr Jason R Vanstone, Saskatchewan Health Authority – Regina Area, 4B35, 1440-14th Avenue, Regina SK S4P 0W5, email:jason.vanstone@saskhealthauthority.ca
(Return to Top)
Funding: None received. (
Return to Text
)
Acknowledgements
The authors would like to thank Dr Jessica Minion and Dr Jeff Betcher for their helpful guidance in the development of this work.
Canadian Journal of Hospital Pharmacy, VOLUME 74, NUMBER 1, Winter 2021


